I am a rising senior majoring in biology. I have loved birds since I was a kid. I knew I wanted to study birds before I knew what an ornithologist was. The existence of birds has always felt impossible to me. The fact that these intelligent, charismatic, diverse, fascinating creatures exist is absolutely astounding. I am incredibly grateful for every opportunity that I have had in ornithology so far. Each experience has felt like a gift.
I am currently assisting on a bioinformatics project, scouring literature for bacteria and viruses that are pathogenic to birds. The collected DNA sequence data will be used to interrogate microbiome data collected by Dr. Forsman from tree swallows and purple martins. While reading these papers (>100), I became more and more interested in pathogens in birds, particularly enteric bacteria. I am currently working an in-depth literature review on Salmonella enterica serovars in birds.
My lab experience is not limited to birds. I have worked in the D4 lab, run by Dr. David Jenkins, for two years. In the D4 lab, I have done work with the highly invasive Giant Apple Snail (Pomacea maculata) as well as work identifying insects collected from mosquito traps. My experience with the D4 lab has led to me being very comfortable in the field and at a microscope. Although adapting to the interminable heat and humidity of Florida took some time, I genuinely enjoy fieldwork.
 I am a co-founder of Knighthawk, the University of Central Florida’s Audubon chapter. I also participated in Audubon’s Conservation Leadership Initiative in 2019, the first year-long program in its history.
I am a co-founder of Knighthawk, the University of Central Florida’s Audubon chapter. I also participated in Audubon’s Conservation Leadership Initiative in 2019, the first year-long program in its history.
Although a lot of my life, both in and out of school, is focused on birds, I have many other interests. I love baking, music (particularly Queen and Freddie Mercury’s solo work), reading, writing, and playing video games.
Julia Nadeau Gneckow CV
NAOC 2020 Poster Presentation (pdf: FinalPoster_Julia)
My name is Julia Nadeau Gneckow and I am a senior at the University of Central Florida.
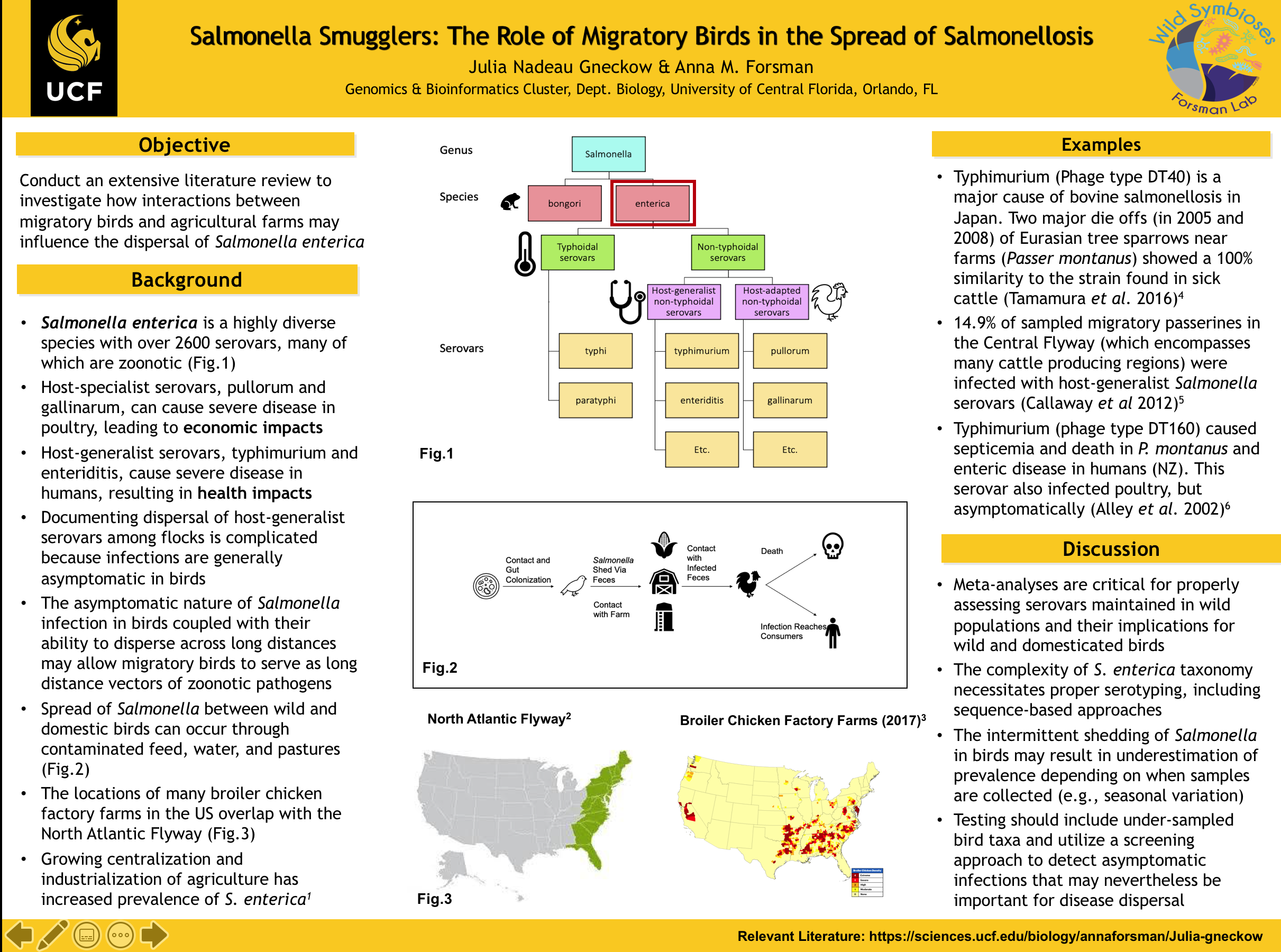
I am currently working on an in-depth literature review of Salmonella in birds. This poster provides an overview of relevant Salmonella taxa and discusses how the dispersal of Salmonella enterica serovars may be influenced by interactions between wild birds and agricultural animals.
As shown in figure 1, there are two species of Salmonella, bongori and enterica. Bongori is typically associated with cold-blooded hosts whereas Enterica infects warm-blooded hosts. Enterica is a highly diverse species with over 2600 serovars, many of which are zoonotic. The serovars can be divided into two major categories, typhoidal and non-typhoidal. Non-typhoidal serovars are the ones we associate with consuming undercooked meat. Host-generalist serovars, such as typhimurium and enteriditis cause gastroenteritis in humans and thus pose significant health concerns. However, infections by these serovars in poultry are generally asymptomatic. Conversely, host-specific serovars like pullorum and gallinarum can cause severe illness, abortion, and death in birds, resulting in economic losses, but they do not generally affect human health. The asymptomatic nature of many Salmonella infections in birds means that dispersal between wild birds and domestic animals, particularly poultry, may go undetected. Wild birds and domestic animals may exchange Salmonella serovars on farms by contaminating feed, water troughs, or pastures.
Salmonella serovars may be picked up and moved large distances by migratory birds. Figure 3 highlights the overlap between broiler chicken factory farms and the North Atlantic Flyway, indicating plenty of opportunities for interaction. Although Salmonella is typically associated with poultry, many of the host-generalist serovars that infect birds can also infect cattle. In fact, many of the beef cattle factory farms in the USA are located along and the Central flyway. In Hokkaido, Japan, eight isolates collected from cows in 2008 were identified as the host-generalist serovar typhimurium (phage type DT40). These cattle infections coincided with a massive die off of Eurasian tree sparrows that were in close proximity to the farm. The Salmonella isolates from the cows and sparrows showed 100% similarity, pointing to bacterial dispersal between these host species.
In another study in North America, passerines captured in close proximity to cattle farms were screened for salmonellosis during migration in 2012. The overall prevalence of Salmonella infection was 14.9%, all of which were asymptomatic infections. Prevalence was also found to increase with proximity to farms. Such asymptomatic infections, coupled with ready access to agricultural farms along migratory routes, may allow migratory birds to serve as long-distance vectors of salmonellosis if more than one farm is visited.
Although rarely documented, mass die-offs of wild birds due to Salmonella present important opportunities for establishing links between humans, agricultural animals, and wild birds. In 2000, Salmonella typhimurium (phage type DT160) was responsible for mass die offs in Eurasian tree sparrows and also increased cases of gastroenteritis in humans in New Zealand. Interestingly, poultry were also infected with this same serovar but presented asymptomatically, demonstrating the potential for undetected dispersal among hosts.
The role of migratory birds in the dispersal of Salmonella continues to be debated in the literature. The complexity and diversity of the Salmonella enterica species complex necessitates proper serotyping in studies to better understand differential infectivity, outcome, and dispersal abilities among serovars. DNA sequencing techniques allow for more specific identification of serovars, which would improve our ability to compare results between studies. Further, utilizing a screening approach to sample all birds captured, regardless of symptoms, will allow us to detect asymptomatic serovars that may be significant to the dispersal of salmonellosis. These are ideas that I will continue exploring during my senior year at UCF.
NAOC 2020 Poster Presentation – References Cited & Relevant Literature
-
MR Alley, JH Connolly, SG Fenwick, GF Mackereth, MJ Leyland, LE Rogers, M Haycock, C Nicol & CEM Reed(2002) An epidemic of salmonellosis caused by Salmonella Typhimurium DT160 in wild birds and humans in New Zealand, New Zealand Veterinary Journal, 50:5, 170-176, DOI: 1080/00480169.2002.36306
-
Figure 3. (October, 2019). Vulnerable Birds in North Atlantic Flyway [Image]. August 1, 2020, from https://www.audubon.org/climate/survivalbydegrees/flyway/atlantic
-
Todd R. Callaway, Tom S. Edrington, and David J. Nisbet. Foodborne Pathogens and Disease. Oct 2014.791-794. http://doi.org/10.1089/fpd.2014.1800
-
Figure 3: Food and Water Watch (May, 2020). BRAND NEW: See America’s Factory Farms Mapped Out [Image]. August 1, 2020, from https://www.foodandwaterwatch.org/news/brand-new-see-americas-factory-farms-mapped-out
-
Gal-Mor O, Boyle EC, Grassl GA. Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front Microbiol. 2014;5:391. Published 2014 Aug 4. doi:10.3389/fmicb.2014.00391
-
Hohmann EL. Nontyphoidal salmonellosis. Clin Infect Dis. 2001;32(2):263-269. doi:10.1086/318457
-
Spickler, Anna Rovid, Leedom Larson, KR. 2013. Salmonellosis. Retrieved from http://www.cfsph.iastate.edu/DiseaseInfo/factsheets.php.
-
Tamamura Y, Uchida I, Tanaka K, et al. A case study on Salmonella enterica serovar Typhimurium at a dairy farm associated with massive sparrow death. Acta Vet Scand. 2016;58:23. Published 2016 Apr 26. doi:10.1186/s13028-016-0205-8